What is the RMSD and how to compute it with GROMACS
The gmx rms command
GROMACS is a powerful open source molecular simulation packages allowing us to run a Molecular Dynamics (MD) simulation, and analyze the resulting trajectories.
Among many tools, at our disposal, we have a specific command to compute the RMSD.
Here I explain to you why the RMSD is an important structural feature in molecular modeling, and how to compute it with GROMACS.
RMSD in Molecular Dynamics
The Root Mean Square Deviation (RMSD) measures how much a certain molecular structure deviates from a reference geometry.
This feature plays a key role in the field of molecular modeling, and you will find it referenced in various papers.
But why is it so important?
In Molecular Dynamics (MD), we are interested in how our system evolves over time as compared to its starting structure. For this reason, plotting the RMSD as a function of time can help us in two ways:
-
First of all, it is useful to identify parts of the simulation where we have important changes in the structure. For example, if a protein over the course of the simulation has some large conformational changes we should notice a spike in the RMSD vs. time plot.
-
As a second point, The RMSD is commonly used as an indicator of convergence of the structure towards an equilibrium state. A flattening of the RMSD curve can indicate that the protein has equilibrated and that we can proceed with the next step.
Therefore, the RMSD has a critical role in the different steps of an MD simulation (i.e., equilibration and production runs).
The equation for RMSD
Let’s discuss the formula needed to compute this fundamental structural feature.
For a general molecular structure with $N$ atoms, we can compute the RMSD between a certain conformation ($r$) and a reference structure ($r^{ref}$) via the following equation:
$$ RMSD = \sqrt{\frac{1}{N}\displaystyle\sum_{i=1}^N(r_i - r_i^{ref})^2} $$
where $r_i^{ref}$ represents the set of coordinates of the $i$-th atom in the reference structure, while $r_i$ is the set of coordinates of the same atom but belonging to the structure that is going to be compared to the reference.
$$ r_i = x_i + y_i + z_i $$ $$ r_i^{ref} = x_i^{ref} + y_i^{ref} + z_i^{ref} $$
In summary, the RMSD is the square root of the average of the squared distances between atoms.
During a simulation, we actually compute the RMSD as a function of time $t$.
At each time step, we compute the RMSD between the coordinates at that specific time step $r(t)$, and the reference.
$$ RMSD(t) = \sqrt{\frac{1}{N}\displaystyle\sum_{i=1}^N(r_i(t) - r_i^{ref})^2} $$
In this way, we can see how the system deviates from the reference structure during our MD simulation.
The gmx rms command
In GROMACS we compute the RMSD via the gmx rms command.
The workflow for this command is quite intuitive. You only need two things:
- A reference geometry in the tpr or gro format
- A trajectory file that is going to be compared to the reference (xtc file)
GROMACS will output a file in the xvg format that you can plot using Grace or Python as I showed in this article .
Here I am going to show you a few examples of how you can play with the gmx rms command to compute the RMSD of your system.
RMSD of a trajectory
First of all, we are going to consider the most basic example.
How can we use the gmx rms command to measure the RMSD fluctuation of the system along the simulation?
In other words, we want to observe how much the structure of the system deviates from the starting one.
In GROMACS this is accomplished by comparing each frame in the trajectory file with the reference structure.
To this aim, you can type the following:
|
|
- Call the program with
gmx - Select the
rmscommand - Select the
-fflag and provide the xtc file from your simulation (simulation.xtc) - Choose the reference structure (
reference.tpr) with the-sflag - Call the
-oflag and decide how you want to name the output xvg file (rmsd.xvg)
- You may want to change the time unit with the
-tu nsoption to output the xvg file in terms of ns instead of ps.
At this stage, you will be prompted to select two groups.
1. The group for least square fits: structure under -f flag
|
|
2. The group for RMSD calculation: structure under -s flag
|
|
For protein simulations, it is a common practice to compute the “backbone to backbone” RMSD value (Select group “4” for both).
You can automate the selection with this command:
|
|
The resulting rmsd.xvg file can be plotted with Grace.
|
|
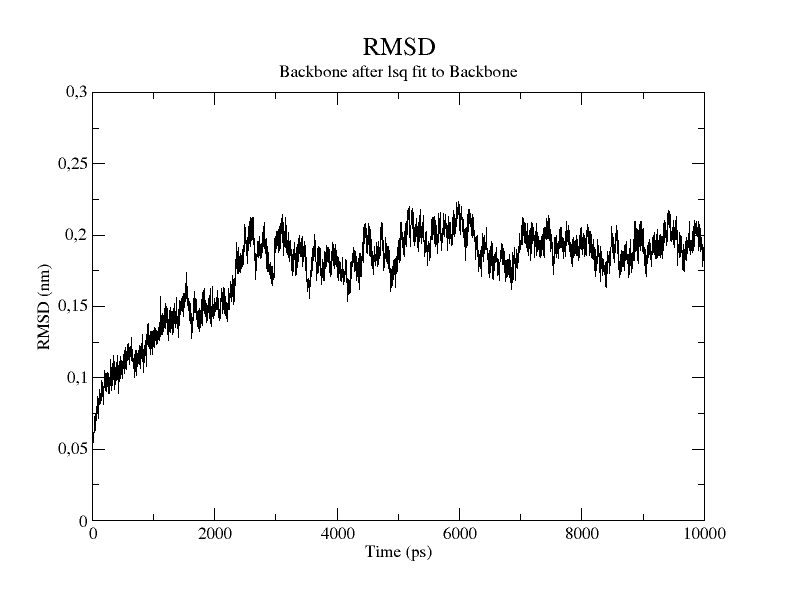
Or with Python using the Numpy and Matplotlib libraries.
|
|

RMSD for a specific part of the system
If you want to analyze a specific part of your system that is not within the default GROMACS groups you can create an index with the gmx make_ndx command.
Subsequently, you can pass the special index that you created to the gmx rms command.
|
|
- Call the program with
gmx - Select the
rmscommand - Select the
-fflag and provide the xtc file from your simulation (simulation.xtc) - Choose the reference structure (
reference.tpr) with the-sflag - Call the
-oflag and decide how you want to name the output xvg file (rmsd_index.xvg) - Prompt the special index you created (
index.ndx) via the-nflag
RMSD for a specific time frame
You can compute the RMSD for a specific time frame of your trajectory by using the -b and -e flags.
For instance, if you are only interested in measuring the RMSD between 1000 ps and 2000 ps of your trajectory you can use the following command.
|
|
- Call the program with
gmx - Select the
rmscommand - Select the
-fflag and provide the xtc file from your simulation (simulation.xtc) - Choose the reference structure (
reference.tpr) with the-sflag - Call the
-oflag and decide how you want to name the output xvg file for that specific time frame (rmsd_time.xvg) - Pick the starting time (1000 ps) for the RMSD calculation using the
-bflag - Select the ending time (2000 ps) using the
-eoption
By selecting the same value for the -b and -e flags you will get the RMSD value between the reference and the frame corresponding to the time you selected.